The Structural Biology and Drug Discovery Shared Resources (SBDDSR) serves as a portal for access to:
- Expertise, consultation and state-of-the-art instrumentation for X-ray crystallography and structure modeling and refinement; and
- Computational resources and expertise to handle the development of structural and force field models, deployment of quantum mechanical calculations, virtual screening of compound libraries, and molecular dynamics studies. Among other computational resources, SBDDSR has access to the supercomputing capabilities of the Wake Forest DEAC cluster. A key feature of SBDDSR is expertise in virtual screening for small molecules, which complements recent high-throughput screening partnerships between SRs at AHWFBCCC and other academic centers in North Carolina.
- Medicinal chemistry expertise to evaluate hits identified from computational and HTS screening efforts including the synthesis of select analogs.
The SBDDSR meets the growing needs for structure determination and computational analysis of protein and DNA/RNA structure, function and dynamics for a diverse array of projects ranging from basic science questions to drug design.
The Structural Biology and Drug Discovery resource supports areas of basic science research with an emphasis on biological processes related to cancer such as:
- Cell signaling
- Transcriptional regulation
- DNA damage and repair
- Lipid metabolism
Basic science research to understand the normal and pathophysiological function of proteins and their interactions with a myriad of biological partners and functional modifiers (e.g., other proteins, DNA, RNA, cofactors, drugs, post-translational modifications, mutations) is the foundation on which to build all progress towards these objectives. We provide access to cutting-edge modeling and simulation methods. The information from these complementary approaches can be used to develop novel therapies.
The SBDD also provides support for ongoing projects and the development of new projects through the collection of preliminary data for funding applications.
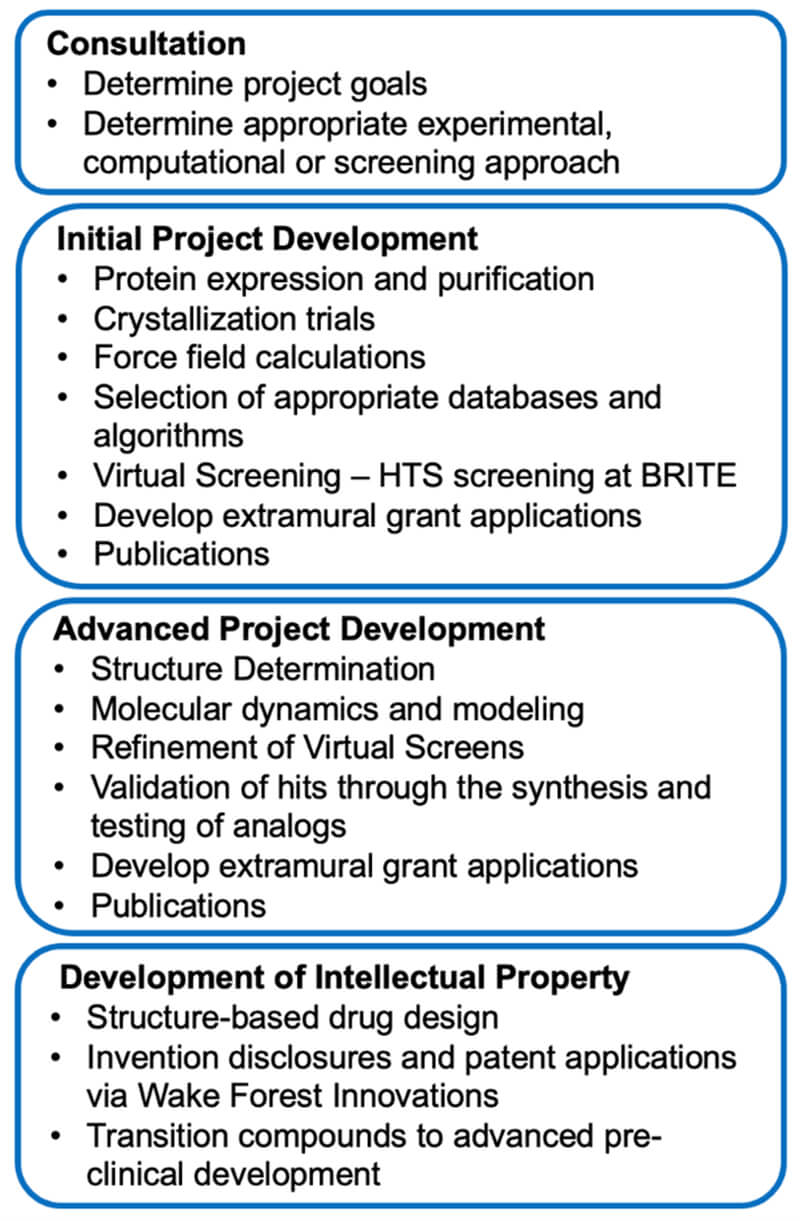
Below is a workflow overview in the SBDDSR: progression from project prioritization and initial development to advanced development and translation.
- Simermeyer TL, Batalis S, Rogers LC, Zalesak OJ, Hollis T. Protein oxidation increases SAMHD1 binding ssDNA via its regulatory site. Nucleic Acids Res 2023;51:7014-24
- Ghoneum A, Gonzalez D, Afify H, Shu J, Hegarty A, Adisa J, et al. Compound C Inhibits Ovarian Cancer Progression via PI3K-AKT-mTOR-NFkappaB Pathway. Cancers (Basel) 2022;14
- Doheny D, Manore S, Sirkisoon SR, Zhu D, Aguayo NR, Harrison A, et al. An FDA-Approved Antifungal, Ketoconazole, and Its Novel Derivative Suppress tGLI1-Mediated Breast Cancer Brain Metastasis by Inhibiting the DNA-Binding Activity of Brain Metastasis-Promoting Transcription Factor tGLI1. Cancers (Basel) 2022;14
- Nelson KJ, Messier T, Milczarek S, Saaman A, Beuschel S, Gandhi U, et al. Unique Cellular and Biochemical Features of Human Mitochondrial Peroxiredoxin 3 Establish the Molecular Basis for Its Specific Reaction with Thiostrepton. Antioxidants (Basel) 2021;10
- Hemphill WO, Simpson SR, Liu M, Salsbury FR, Jr., Hollis T, Grayson JM, et al. TREX1 as a Novel Immunotherapeutic Target. Front Immunol 2021;12:660184
- Booth WT, Clodfelter JE, Leone-Kabler S, Hughes EK, Eldeeb K, Howlett AC, et al. Cannabinoid receptor interacting protein 1a interacts with myristoylated Galpha(i) N terminus via a unique gapped beta-barrel structure. J Biol Chem 2021;297:101099
- Bianchi-Smiraglia A, Wolff DW, Marston DJ, Deng Z, Han Z, Moparthy S, et al. Regulation of local GTP availability controls RAC1 activity and cell invasion. Nat Commun 2021;12:6091
- Batalis S, Rogers LC, Hemphill WO, Mauney CH, Ornelles DA, Hollis T. SAMHD1 Phosphorylation at T592 Regulates Cellular Localization and S-phase Progression. Front Mol Biosci 2021;8:724870







